Supplementary Information for an Experiment on:
Beta-Galactosidase in Escherichia coli
in the UW-Madison Bacteriology 304 Course
Fall Semester, 2003 and Summer Session, 2007
|
|
I. A General Summary of the Lac Operon.
The movement of RNA Polymerase along the lac operon (i.e., the direction of transcription) is from left to right.
|
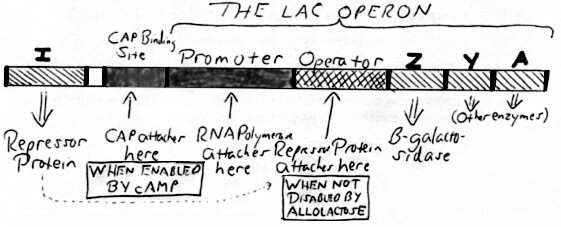
- RNA Polymerase Attachment: The Catabolic Activator Protein (CAP) attaches to the CAP BINDING SITE (when CAP is enabled by cAMP) and allows RNA Polymerase to attach to the PROMOTER.
- RNA Polymerase Transcription of the Lac Operon then proceeds – unless blocked by the Repressor Protein (RP) which attaches to the OPERATOR (when RP is not disabled by allolactose).
|
|
II. A Review of Activation and Repression of the Lac Operon
based on factors which allow (or block) attachment of – and transcription by – RNA polymerase.
| |
GLUCOSE
cAMP
CAP enabled by cAMP to allow attachment of RNA polymerase.
|
GLUCOSE
cAMP
CAP disabled, so RNA polymerase cannot attach.
|
|
LACTOSE
Allolactose * *
RP disabled by allolactose, allowing transcription of lac operon to proceed (if RNA polymerase can attach).
|
RNA polymerase:
- can attach,
- and can transcribe lac operon (as RP blockage is disabled).
IDEAL SITUATION
|
RNA polymerase:
- cannot attach;
- therefore, no transcription of the lac operon (even though there is no blockage by RP).
|
|
LACTOSE
Allolactose
Repressor Protein (RP) is free to block lac operon transcription.
|
RNA polymerase:
- can attach,
- but cannot transcribe (due to blockage by RP).
|
RNA polymerase:
- cannot attach;
- therefore, no transcription of the lac operon (which would be blocked by RP anyway).
|
*Basal enzymatic activity is always present which allows the initial production of allolactose when lactose is introduced into the system. (Even though no beta-galactosidase production is expected when glucose is present, there is still a low level of transcription.)
 A question from a Bacteriology 304 quiz given August, 2007: What should theoretically happen when a typical strain of Escherichia coli is inoculated into a medium containing high amounts of both glucose and lactose? The two-part answer:
A question from a Bacteriology 304 quiz given August, 2007: What should theoretically happen when a typical strain of Escherichia coli is inoculated into a medium containing high amounts of both glucose and lactose? The two-part answer:
- Elevated glucose causes a decrease in cAMP. With less or no cAMP, the Catabolic Activator Protein (CAP) is disabled. Therefore there is no attachment of the RNA polymerase and thus no transcription of the lac operon. This way, the glucose is used preferentially to the lactose as a carbon and energy source.
- When the glucose runs out, the increase in cAMP enables the CAP to attach, allowing the RNA polymerase to attach and proceed with transcription of the lac operon which – with the elevated lactose present – is not blocked by the repressor protein (RP).
Application of the Boolean circuit idea to the lac operon is discussed here.
III. A Sample Calculation.
A flow chart for a timed beta-galactosidase activity experiment is shown here. The "assigned culture" may be a wild-type or mutant strain in a standard broth medium containing glucose, lactose or both. (Why not see what happens in a galactose broth.)
The following is an example of a calculation to determine units of beta-galactosidase per ml of a normalized culture in a timed experiment where we are testing (with a spectrophotometer set at 420nm) a 4 ml assay tube composed of 1 ml of culture, 1 ml of buffer, 1 drop of toluene (an insignificant amt.), 1 ml of ONPG, and 1 ml of stop solution.
Comparison is made with a cell absorbance reading (determined at 600nm) of the same culture at the same time of testing for the normalization of the result (i.e., based on constant cell density).
For this example calculation we have the following data that are plugged into the sequential formulas:
- "e" (the extinction coefficient): 1.1 X 103 ml/(micromole X cm)
- "l" (the length of the light path): 1 cm
- A600 of the culture: 0.220
- A420 of the assay: 0.346
- Time of the assay: 1.33 min

IV. Some Data for Graphing Practice.
One may get an impression of the nature of the mutant strain – that is, what mutation(s) may have allowed the results shown?
| strain & medium |
time (min) |
A420 |
A600 |
time of
assay (min) |
U/ml |
U/(ml X A600) |
| mutant strain in MM+glucose |
0 |
0.033 |
0.03 |
8.33 |
1.44E-05 |
4.80E-04 |
| 15 |
0.34 |
0.05 |
7.033 |
1.76E-05 |
3.52E-04 |
| 30 |
0.34 |
0.04 |
6.67 |
1.86E-05 |
4.64E-04 |
| 45 |
0.04 |
0.05 |
7.0 |
2.08E-05 |
4.16E-04 |
| 60 |
0.066 |
0.06 |
7.0 |
3.43E-05 |
5.71E-04 |
| 75 |
0.109 |
0.12 |
7.033 |
5.64E-05 |
4.70E-04 |
| 90 |
0.24 |
0.28 |
5.33 |
1.64E-04 |
5.85E-04 |
| 105 |
0.246 |
0.24 |
5.5 |
1.63E-04 |
6.78E-04 |
| 120 |
– |
– |
– |
– |
– |
| mutant strain in MM+lactose |
0 |
0 |
0.009 |
10.0 |
0 |
0 |
| 15 |
0.035 |
0.246 |
10.0 |
1.27E-05 |
5.17E-05 |
| 30 |
0.055 |
0.303 |
10.0 |
2.00E-05 |
6.60E-05 |
| 45 |
0.198 |
0.376 |
8.5 |
8.47E-05 |
2.25E-04 |
| 60 |
0.207 |
0.468 |
4.53 |
1.66E-04 |
3.55E-04 |
| 75 |
0.247 |
0.562 |
4.33 |
2.07E-04 |
3.69E-04 |
| 90 |
0.349 |
0.608 |
2.83 |
4.48E-04 |
7.38E-04 |
| 105 |
0.85 |
0.756 |
1.69 |
1.83E-03 |
2.42E-03 |
| 120 |
– |
– |
– |
– |
– |
| wild-type strain in MM+glucose |
0 |
0 |
0.03 |
5.0 |
3.64E-06 |
1.21E-04 |
| 15 |
0.015 |
0.078 |
5.0 |
1.09E-05 |
1.40E-04 |
| 30 |
0.008 |
0.08 |
5.0 |
5.82E-06 |
7.27E-05 |
| 45 |
0.02 |
0.17 |
5.0 |
1.45E-05 |
8.56E-05 |
| 60 |
0.021 |
0.18 |
5.0 |
1.53E-05 |
8.48E-05 |
| 75 |
0.024 |
0.26 |
5.0 |
1.75E-05 |
6.71E-05 |
| 90 |
0.019 |
0.4 |
5.0 |
1.38E-05 |
3.45E-05 |
| 105 |
0.016 |
0.49 |
5.0 |
1.16E-05 |
2.37E-05 |
| 120 |
– |
– |
– |
– |
– |
| wild-type strain in MM+lactose |
0 |
0 |
0 |
10.0 |
0 |
0 |
| 15 |
0 |
0.07 |
10.0 |
0 |
0 |
| 30 |
0.038 |
0.03 |
10.0 |
1.38E-05 |
4.61E-04 |
| 45 |
0.024 |
0.04 |
10.0 |
8.73E-06 |
2.18E-04 |
| 60 |
0.078 |
0.06 |
10.0 |
2.84E-05 |
4.73E-04 |
| 75 |
0.21 |
0.07 |
10.0 |
7.64E-05 |
1.10E-03 |
| 90 |
0.247 |
0.14 |
10.0 |
8.98E-05 |
6.42E-04 |
| 105 |
– |
– |
– |
– |
– |
| 120 |
– |
– |
– |
– |
– |
|
Page last modified on 1/10/17 at 1:00 PM, CST.
John Lindquist, Department of Bacteriology
University of Wisconsin – Madison
|
|
I can be e-mailed at
jlindquist 001 @ gmail.com
(Use no spaces.)
|



 A question from a Bacteriology 304 quiz given August, 2007: What should theoretically happen when a typical strain of Escherichia coli is inoculated into a medium containing high amounts of both glucose and lactose? The two-part answer:
A question from a Bacteriology 304 quiz given August, 2007: What should theoretically happen when a typical strain of Escherichia coli is inoculated into a medium containing high amounts of both glucose and lactose? The two-part answer: