Microbiology 102 Experiment 8:
|
|
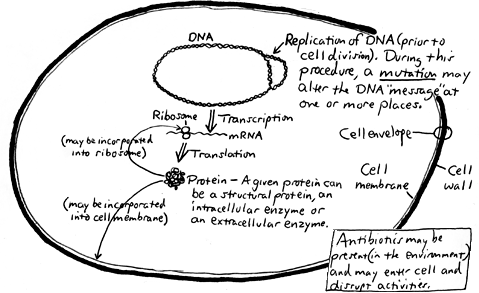
Here is a simplified scheme of a typical bacterial cell showing DNA replication, transcription and translation processes which occur simultaneously and continuously:
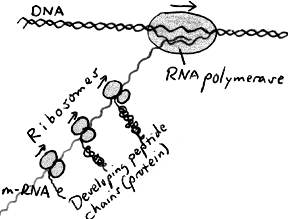
The diagram on the right shows transcription and translation in a little more detail.
DNA may be changed by mutation or recombination. When this happens, the phenotype may be altered (i.e., a change in a visible characteristic may be seen), or the cell may die due to a lack of a needed function. MUTATIONA mutation is a spontaneous change in the DNA sequence which is passed on to the "offspring." This change, generally occuring during DNA replication, may be accomplished by substitution, deletion or insertion of one or more bases such that the base sequence is altered, leading to other than the intended protein being constructed. Most mutations are deleterious, leading to death of the cell. Those mutations which are not lethal may cause a visible alteration of the phenotype and may confer a special survival benefit to the organism, possibly allowing the organism to colonize a previously-hostile environment. A mutation may lead to one or more of the following changes in the phenotype of the cell:
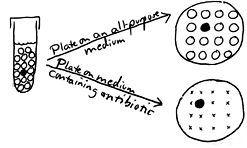
The following over-simplified example shows selection of antibiotic-resistant cells from a culture of a species generally thought of as being sensitive to the antibiotic. A test tube culture containing sixteen cells is shown on the left in this diagram. All of the cells are sensitive to a particular antibiotic except for one cell which because of a mutation is resistant. If the culture is dumped onto a plate of an all-purpose medium, all of the cells will form colonies. One cannot distinguish colonies of antibiotic-resistant cells from colonies of antibiotic-sensitive cells. If the culture is dumped instead onto a plate of the same medium to which is added the antibiotic, the sensitive cells will be inhibited or killed by the antibiotic the selective agent in the medium while the resistant cell will form a colony. A take-home lesson: Suppose that this test tube is your body and the bacterial cells are infecting it. If you are given this antibiotic to control the infection, will it be totally effective? Might the resistant mutant escape your body's own natural defenses and then multiply and continue the infection? This is an important way in which antibiotic-resistant strains of a pathogen can arise. In the antibiotic disk sensitivity test (one of our virtual experiments), we test various species of bacteria (potential pathogens) with a number of different antibiotics, and we can determine the best antibiotics to use against a given organism where there is no indication of any growth of resistant cells. |
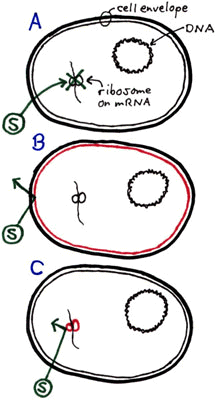
The antibiotic streptomycin intereres with ribosomes in "streptomycin-sensitive" cells, stopping translation. The ribosome is the target site for streptomycin. In the diagram on the right, cell A represents a typical Staphylococcus epidermidis cell (somewhat flattened, however) that is sensitive to streptomycin (represented by the circled S) which is able to get through the cell membrane and stop protein synthesis at site of the ribosome. Cell B represents a cell whose membrane is altered by a mutation such that streptomycin no longer has easy access to the interior of the cell. Colonies having this type of mutation tend to be considerably smaller than normal-sized S. epidermidis colonies. Why might this be so? Would we expect an increase of the concentration of streptomycin to be more inhibitory to this type of mutant? Cell C represents a cell whose ribosome is altered by a mutation such that streptomycin no longer is able to stop protein synthesis. The cell carries on and produces colonies of normal size. no matter what concentration of streptomycin we used. Would we expect an increase of the concentration of streptomycin to be more inhibitory to this type of mutant? The percentage of mutants in a population can be determined. As an example, the mutation frequency in the above test tube culture of 16 cells is 1 out of 16 or 6.25%. (However, would be an unreasonable example, given the fact that there is about one chance in a million that a given gene will mutate when the DNA is replicated.) |
CONJUGATION AND RECOMBINATION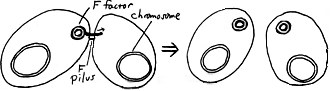
In Escherichia coli and presumably many other species, distinct "mating types" may be found. For E. coli, the F+ and F mating types are distinguished by the presence of a special plasmid, the F factor, in the former. This plasmid includes genes involved in the transfer by conjugation of DNA into compatible recipient cells; included are genes responsible for the synthesis of the F pilus. As shown in this diagram, when the F plasmid is transferred to an F cell, the donor F+ cell retains a complete copy of the plasmid while the recipient F cell becomes an F+ donor cell. |
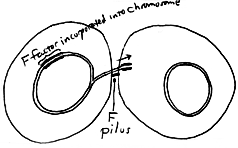
If the F plasmid becomes integrated into the chromosome of a F+ cell, an Hfr cell results. A cell of this mating type will be a donor, able to transfer part of its regular chromosome during conjugation (with part of the F factor on the leading end). Insertion of any part of the transferred Hfr chromosome into the recipient cell's chromosome (recombination) may subsequently occur. In order for us to demonstrate the result of conjugation and recombination effectively in this experiment, we must choose parent cells which are not only different mating types but are also different in one or more phenotypic properties such that we may detect the result of recombination easily. That is, we should see some kind of growth or reaction occuring after these two parent cultures are mixed that we do not see for the two separate parent cultures. That is the take-home lesson from this experiment. We have chosen an Hfr strain which is a methionine auxotroph and an F strain which is a threonine auxotroph. Neither should grow on a minimal medium which is formulated for prototrophic ("wild-type") strains of E. coli. However, after mixing the two strains and allowing for conjugation and subsequent recombination to occur, the appearance of any colony on the minimal medium can be attributed to an F cell which acquired the ability, from an Hfr cell (via conjugation and recombination), to synthesize threonine and thus all of its amino acids. The following table summarizes the important characteristics of our strains:
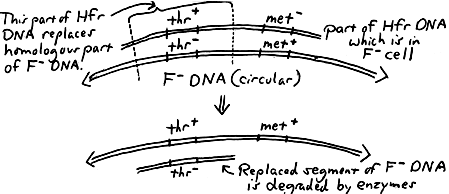
The following diagram shows an overview of the "mechanics" of recombination leading to the recipient (F) cell becoming thr+ that is, gaining the ability to synthesize threonine from the inclusion of that part of the donor DNA into its chromosome. The cell is thus able to (1) produce all of its amino acids and (2) form a colony on Minimal Medium:
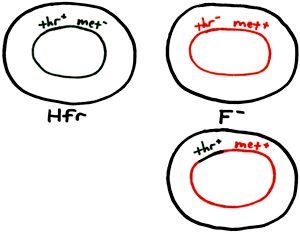
Another way of looking at it (with a "before and after" view of the F cell) is shown on the right. As an example of what to consider in the calculation of recombination frequency, here is a sample question from a quiz: Ten Hfr cells are mixed with ten F cells, and four pairs of Hfr and F cells undergo conjugation. If one F cell incorporates DNA from an Hfr cell into its chromosome, the "recombination frequency" would then be . (The answer would not be 1 out of 20 (0.5%) but rather 1 out of 10 (10%). Consider that only the F cells are able to undergo recombination, and the number of Hfr cells should not be considered in the calculation. More realistically, we usually find the recombination frequency to be between 0.05 and 0.1%.) |